Author Chris Janse
- Genome and genome sequence
- Genome size, base composition, mitochondrial and plastid DNA, DNA replication
- Chromosomes, centromeres, telomeres, subtelomeric and core regions
- Chromosome size variation, DNA-rearrangements
Genome and genome sequence
See for detailed information on the genome and genome sequence of P. berghei (and other rodent malaria parasites) the following papers:
- Hall et al., (2005). A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005 Jan 7;307(5706):82-6.
- Otto et al. (2014). A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014 Oct 30;12:86.
Table: Features of the genomes of rodent malaria parasites (and compared with the human parasite P. falciparum) (from Otto et al. (2014). BMC Biol. 2014 Oct 30;12:86).
| Genome features | P. berghei ANKA | P. c. chabaudi AS | P. y. yoelii (YM) | P. falciparuma |
| Nuclear genome | ||||
| Genome size (Mb) | 18.5 | 18.8 | 21.9 | 23.3 |
| G+C content (%) | 22.1 | 23.6 | 21.1 | 19.4 |
| Chromosomes | 14 | 14 | 14 | 14 |
| Synteny breaksb | – | 1 | 0 | ND |
| Contigs | 220 | 40 | 195 | 14 |
| Sequence coverage | 237x | 109x | 627x | – |
| Genesc | 4,979 | 5,139 | 5,675 | 5,419 |
| Genes with functional annotationd | 2,781 (56%) | 2,927 (57%) | 3,485 (61%) | 3,234 (60%) |
| Mitochondrial genome | ||||
| Genome size (bp) | 5,957 | 5,949 | 6,512 | 5,967 |
| G+C content (%) | 30.9 | 30.9 | 30.7 | 31.6 |
| Number of genes | 3 | 3 | 3 | 3 |
| Apicoplast genome | ||||
| Genome size (bp) | 30,302 | 29,468 | 29,736 | 29,430 |
| G+C content (%) | 13.5 | 13.7 | 14.1 | 13.1 |
| Number of genes | 30 | 30 | 30 | 30 |
Genome size, base composition, mitochondrial and plastid DNA, DNA replication
Genome size
The genome is organised into 14 chromosomes and has a total size of 18.5 Mb.
Base composition
Like P. falciparum, the nuclear DNA of P. berghei has a high overall A+T content of about 80%. This (A+T)-rich bias is unevenly distributed between protein-coding and non-coding regions. All open reading frames are relatively (G+C)-rich (25-30%), while the (A+T) composition of the vast majority of the intergenic regions and intragenic introns can rise to more than 90%.
Extranuclear DNA: mitochondrial and plastid genome
P. berghei has two extra-nuclear DNA elements comparable to P. falciparum: the mitochondrial DNA and the plastid DNA (organelle genomes). All Plasmodium species analysed so far contain a ~6 kb tandemly repeated mitochondrial (mt) genome which codes for only three proteins (cytochrome b and two subunits of cytochrome oxidase) as well as two fragmented rRNA’s. The circular plastid genome (apicoplast) is ~30 kb in size. The apicoplast is a vestigial plastid homologous to the chloroplasts of algae and plants. The plastid (known as the apicoplast; for apicomplexan plastid) is non-photosynthetic and very much reduced, but has clear endosymbiotic ancestry including a circular genome that encodes RNAs and proteins and an ensemble of bacteria-like pathways to replicate and express its genome plus an anabolic capacity generating fatty acids, haem and isoprenoid precursors. A key process within the apicoplast is the synthesis of the five-carbon isoprenoid precursor molecules. All isoprenoids are derived from these precursors and isoprenoid functions are required in all living cells. These molecules fulfill a variety of cellular roles, including participation in key processes such as N-glycosylation, electron transport (ubiquinone), and protein prenylation. Isoprenoid biosynthesis occurs via a metabolic pathway, known as the methylerythritol phosphate (MEP) pathway. Because this organelle is cyanobacterial in origin, the MEP pathway is shared by the majority of eubacteria and other plastid-containing eukaryotes, such as plants and algae.
DNA synthesis and ploidy of life cycle stages
Merozoites, gametes and sporozoites are haploid. The only diploid stage is the ‘young’ zygote, just after fertilization. The dividing stages, such as schizonts and oocysts are ‘polyploid’, because DNA replication and nuclear division are not immediately followed by cell division, resulting in a ‘syncytial’ cell with many nuclei. Only towards the end of schizogony/sporogony does the parasite start to divide its cytoplasm by budding of uninuclear haploid merozoites/sporozoites. The ookinete stage has a nucleus containing the ‘tetraploid’ amount of DNA resulting from fertilization and meiosis without immediate nuclear division. Nuclear division only starts in the oocyst stage.
During asexual blood-stage development DNA synthesis starts around 16 hours after invasion in the old trophozoite, just before the first nuclear division in the schizont. Throughout schizogony DNA replication and genome segregation are alternating events. The timing and rate of DNA synthesis in blood-stages of P. berghei is comparable to that in P. falciparum blood stages, where DNA synthesis also starts in the old trophozoites and continues during schizogony.
Replication of the (extra-nuclear) plastid genome of P. berghei occurs (just before and) during schizogony as has been found in P. falciparum.
During male gametogenesis (formation of male gametes), three rounds of genome replication take place within 10 minutes after activation of the male gametocytes. The content of the resulting ‘octoploid’ nucleus is divided over the eight male gametes, resulting in the haploid male gametes. It has been calculated that the entire haploid genome is replicated in, on average, 3.2 min during the formation of the male gametes.
In the diploid zygotes of P. berghei, DNA synthesis (up to the tetraploid value) coincides with meiotic division. This suggests that this DNA synthesis represents the genome replication during the first meiotic division like in other eukaryotes.
Chromosomes: centromeres, telomeres, subtelomeric and core-regions
Chromosomes
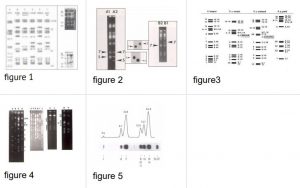
See figures 1-5 for pictures of the 14 chromosomes separated by pulsed field gel electrophoresis.
The chromosomes of Plasmodium species do not condense prior to mitosis like in most other eukaryotes. Due to the lack of condensation and the small size chromosomes cannot be visualized by light-microscopy or by standard electron microscopy. Pulsed field separation of chromosomes revealed that P. berghei has 14 chromosomes in the size range of 0.5-4 Mb (see Figs. 1-5).
Table: The size of the 14 chromosomes of P. berghei (ANKA strain, clone 8417)
| Chromosome Number | Estimated size (Mb) | Presence of 2.3 kb repeats1 | Remarks |
| 13/14 | 3.8 | yes | 13 and 14 co-migrate in most lines; in several clones size differences exist which allows separation by pulsed field gel electrophoresis |
| 12 | 1.9 | yes | P. chabaudi clones exist in which chr. 9, 10 and 11 can be separated, allowing determination of chromosome location of genes |
| 9/10/11 | 1.8 | yes | |
| 8 | 1.6 | no? | |
| 7 | 1.5 | yes | Size is highly variable as a result of loss and acquisition of 2.3kb repeats. Chr. 7 may have the same size or can be smaller than chr. 5/6 (see Fig. 1) |
| 6 | 1.3 | yes | Size is also variable (comparable to chr. 7); therefore not always separated from chr. 5 or 7 (see Fig. 1) |
| 5 | 1.2 | no | |
| 4 | 0.8 | no | |
| 3 | 0.75 | no | |
| 2 | 0.7 | no | |
| 1 | 0.65 | yes |
1Presence of subtelomeric 2.3kb repeat units as shown by hybridisation experiments
Figure: Images and descriptions of chromosomes, chromosome size variation, karyotypes and presence of 2.3 kb subtelomeric repeats.
See PDF-file for larger images
Centromeres and telomeres
Centromeres and telomeres have been (functionally) characterized in P. berghei. Telomeres with the repeat sequence of CCCTA(G)AA have been characterized and this sequence is similar in all Plasmodium species analysed so far. The total length of a telomere is about 1-1.2kb.
Subtelomeric regions
Like the subtelomeric regions of P. falciparum chromosomes, these regions of P. berghei chromosomes contain many (different), non-coding subtelomeric repeat sequences that are species specific. A widely distributed subtelomeric repeat- sequence in P. berghei is the 2.3 kb repeat unit. These units are directly joined to the telomeric repeats of several chromosomes (see table 1). These repeats and variations in copy number have been characterized in detail.
The genomes of rodent malaria parasites contain a number of multigene families located in the subtelomeric chromosomal regions. These include a large family of so-called `Plasmodium interspersed repeat genes’ (pir), that are present also in other human/primate Plasmodium species, such as the human parasite P. vivax. Most of these gene families are expressed in blood stages and these proteins show features that have been reported to contribute to immune evasion through antigenic variation and may play a role in the sequestration of infected red blood cells and virulence.
Table: Different (subtelomeric) multigene families in the genomes of rodent malaria parasite (RMP)
| Gene family (new name) | Other (previous) names | No. of genes | ||||||||
| PbA | PbA | PbA | PcAS | PcAS | PcAS | PyYM | PyYM | PyYM | ||
| CG | PSG | FG | CG | PSG | FG | CG | PSG | FG | ||
| pir | pir, bir, cir, yir | 100 | 88 | 12 | 194 | 3 | 4 | 583 | 40 | 172 |
| RMP-fam-a | Pb-fam-1; Pc-fam-1; fam-a; PYSTA | 23 | 16 | 3 | 132 | 2 | 0 | 94 | 8 | 11 |
| RMP-fam-b | Pb-fam-3; PYSTB | 34 | 1 | 5 | 26 | 0 | 0 | 48 | 2 | 4 |
| RMP-fam-c | PYSTC | 6 | 0 | – | 10 | 0 | – | 22 | 0 | – |
| RMP-fam-d | Pc-fam | 1 | 0 | – | 17 | 4 | – | 5 | 0 | – |
| Early transcribed membrane protein | etramp | 7 | – | – | 13 | – | – | 12 | – | – |
| Reticulocyte binding protein, putative | P235; 235kDA protein rhoptry protein, putative | 6 | – | 8 | 8 | – | 0 | 11 | – | 3 |
| Lysophospholipase | 4 | 1 | – | 28 | 0 | – | 11 | 1 | – | |
| RMP-erythrocyte membrane antigen (RMP-EMA1) | pcema1 | 1 | 0 | 0 | 13 | 1 | 1 | 1 | 0 | 0 |
| haloacid dehalogenase-like hydrolase, putative | 1 | – | – | 9 | – | – | 1 | – | – | |
| ‘Other subtelomeric genes’ | 46 | – | – | 67 | – | – | 46 | – | – |
PbA = P. berghei ANKA, PcAS = P. c. chabaudi AS, PyYM = P. y. yoelii YM. CG: complete gene; FG: fragment; PSG: pseudogene. (from Otto et al. (2014). BMC Biol. 2014 Oct 30;12:86).
Core regions of chromosomes
In comparison with the highly variable subtelomeric regions of Plasmodium chromosomes, the central, core regions are much more stable and conserved between different rodent malaria parasite species. A high level of conservation of gene linkage groups (synteny: gene location and order on chromosomes) exists between the four rodent malaria parasite species rodent parasites, emphasizing a low frequency of large-scale rearrangements in the core regions of their chromosomes. Although the level of synteny of genes is lower when the genomes of rodent and human species of Plasmodium are compared, significant conservation of genome organization has been observed.
A comparison was made of all predicted rodent malaria parasite protein-coding genes with those of three primate malaria species, P. falciparum, P. knowlesi and P. vivax using OrthoMCL where the predicted rodent malaria parasite proteome was divided into three different categories: (1) rodent malaria parasite proteins with orthologs in any of the primate malarias; (2) rodent malaria parasite-specific proteins with no orthologs in primate malarias; and (3) primate malaria-specific proteins with no orthologs in any of the RMP. Between the predicted RMP proteomes (15,793 proteins in total) and primate malaria proteomes (15,853 proteins in total), approximately 87% of the rodent malaria parasite proteins had detectable orthologs in at least one of the primate malarias and only 2,104 proteins (13.3%) were predicted to be rodent malaria parasite-specific. Of those 2,104 proteins, 1,854 (88.1%) are from multigene families. For 2,306 primate malaria proteins (14.6%) no orthologs have been detected in the rodent malaria parasites. Of these primate malaria-specific genes, approximately 1,635 (70.9%) are subtelomeric genes or members of subtelomeric gene families (from Otto et al. (2014). BMC Biol. 2014 Oct 30;12:86).
Chromosome size variation, DNA-rearrangements
See also figures 1 to 5
Size differences between homologous chromosomes of up to 0.5Mb have been detected in parasites from different strains or clones of P. berghei . Size differences result from (large-scale) chromosomal rearrangements, mainly affecting the subtelomeric regions. At this moment no direct evidence exists for the presence of developmentally regulated or programmed DNA rearrangements in Plasmodium by which parasites increase antigenic variation or regulate gene transcription. Most size polymorphisms of chromosomes result from ‘aberrant’ chromosomal rearrangements. These DNA rearrangements occur frequently in the subtelomeric regions, while the internal parts of the chromosomes (core regions) appear to be more conserved. Since many multi-gene families that are associated with antigenic variation are located in the subtelomeric regions, these rearrangements may constitute a significant form of genetic variation. A number of chromosomal rearrangements leading to significant changes in the size of chromosomes have been characterized in P. berghei, including chromosome breakage and healing, loss and acquisition of subtelomeric repeats and chromosome translocation. The mechanisms underlying chromosome size polymorphisms in P. berghei are comparable to those of P. falciparum. In P. berghei chromosome size polymorphisms are most frequently the result of loss and acquisition of subtelomeric repeat sequences. In rodent parasites kept under laboratory conditions, large-scale rearrangements have even been shown to affect chromosome number. In response to drug pressure using the antifolate drug pyrimethamine, part of chromosome 7 containing the drug-sensitive dihydrofolate reductase-thymidylate synthase gene was duplicated to form a small ‘mini-chromosome’ of 400-500kb. This (partial) chromosome duplication, found both in P. chabaudi and P. berghei resulted in a population of parasites with 15 instead of 14 chromosomes.
Research on the mechanisms of chromosome size variation in P. berghei has provided evidence that recombination can occur between non-homologous chromosomes. These kinds of recombination events are significant since they can result in genetic exchange, changing gene location and the clustering of genes.
Table: Chromosomal rearrangements described in P. berghei and in P. falciparum
| (large scale) rearrangements | P. berghei | P. falciparum | |
| Deletion of subtelomeric repeat sequences | yes | yes | |
| Deletion of subtelomeric located genes | yes | yes | |
| Increase in number of subtelomeric repeats | yes | ||
| Acquisition of subtelomeric repeats by recombination between non-homologous chromosomes | yes | yes? | |
| Chromosome translocation | yes | yes | |
| Gene duplication | yes | yes | |
| Chromosome breakage followed by healing through the addition of telomeric sequences | yes | yes | |
| Chromosome duplication | yes | no | |
| Programmed rearrangements resulting in changes in gene expression | no | no |